Accurately model the complexities of human Metabolic dysfunction-associated steatohepatitis (MASH) in vitro
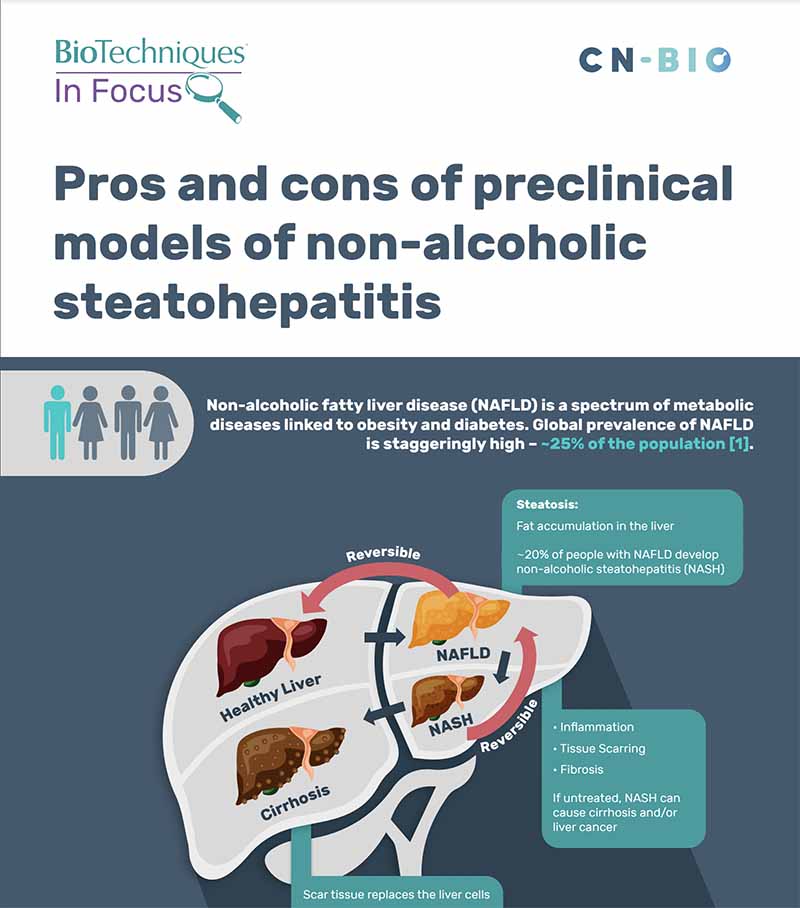
A common cause of liver disease worldwide is Metabolic dysfunction-associated steatotic liver disease (MASLD), 20% of people diagnosed with MASLD develop MASH*, a chronic liver disease that is caused by inflammation from fat buildup in the liver. Left untreated, MASH may progress into cirrhosis and/or liver cancer.
Despite its prevalence, this disease has only one regulatory-approved therapeutic; why is this? MASLD/MASH arises from multiple factors that result in phenotypes that are difficult to replicate in the laboratory. The inability of current preclinical models to replicate human MASH is a contributing factor to drug attrition.
*previously known as Non-Alcoholic Fatty Liver Disease (NAFLD) and Non-alcoholic Steatohepatitis (NASH).
Our solution
The PhysioMimix MASH Assay utilizes a tri-culture Liver-on-a-chip model comprised of human primary liver cells grown in 3D, under perfusion to recapitulate the liver’s physiological environment.
Primary human hepatocytes, Kupffer cells (inflammatory response), and stellate cells (fibrotic response) cultures are treated with proprietary HEP-Fat media to create an in vitro MASH phenotype that closely mimics the key aspects of the disease.
This advanced assay delivers long-term 3D liver microtissue cultures that enable the assessment of therapeutic efficacy or the exploration of disease pathways for target identification. We utilize validated cells with known genotypes, allowing MASH-related single nucleotide polymorphisms (SNPs) to be explored.
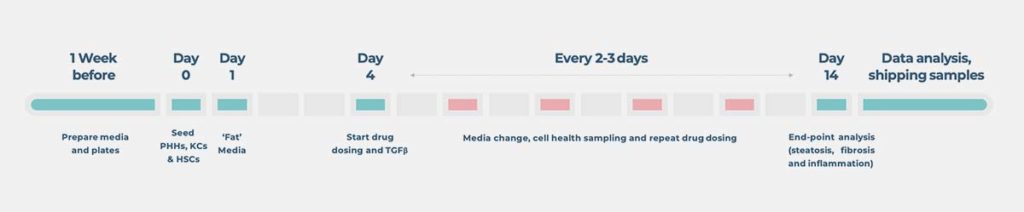
The MASH assay is functional for at least 14 days. It enables the quantification of biomarkers (such as inflammation, fibrosis, and steatosis) and cell health markers (including ALT and AST), allowing translatability to the clinic.
Studying MASLD/MASH
Limitations of current techniques
- Lack of human-relevant preclinical models that capture disease complexity
- Animal models failing to predict clinical outcomes
- Poor translatability of preclinical data
- Short-lived hepatic function in standard cultures limits experiments
- Difficulty tailoring models to suit specific scientific questions
Advancements with PhysioMimix OOC®
- PhysioMimix MASH model replicates key phenotypic biomarkers
- Generate data using key hepatic human cell types to support in vivo findings
- Measure key clinical markers such as ALT and AST
- Phenotypic MASH cultures for at least 14 days to enable a longer-term analysis
- Highly adaptable to specific aspects of the disease to tailor the application to suit your needs
MASLD/MASH infographic
View our “Pros and cons of preclinical models of Metabolic dysfunction-associated steatohepatitis infographic”
End point measurements
Longitudinal and endpoint measurements include (but not limited to):
Functionality biomarkers
- Albumin production
- Urea production
Clinical liver health biomarkers
- Lactose dehydrogenase (LDH) release
- Aspartate Transferase (AST)/Alanine amino transferase (ALT)
Disease biomarkers
- Luminex®/ELISA assays
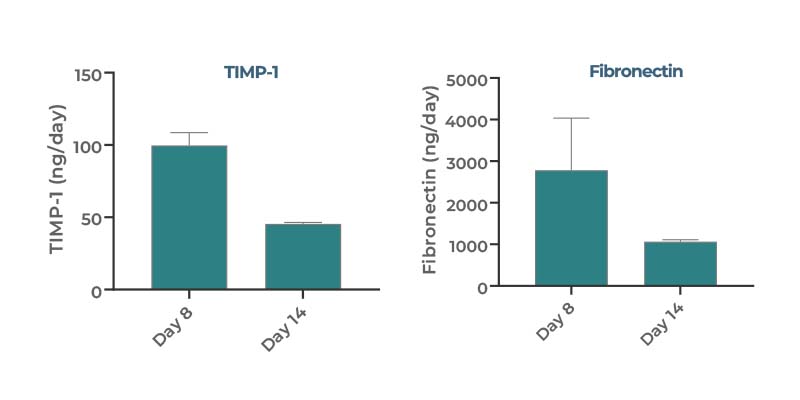
- Fibrosis (e.g. TIMP-1, Pro-collagen, Fibronectin)
- Inflammation (e.g. IL-6, IL-8 TNF-α)
- Confocal microscopy
- Smooth muscle actin
- Collagen
- Fat accumulation (Nile Red staining)
Optional profiling analysis
- Quantitative PCR
- Transcriptomics
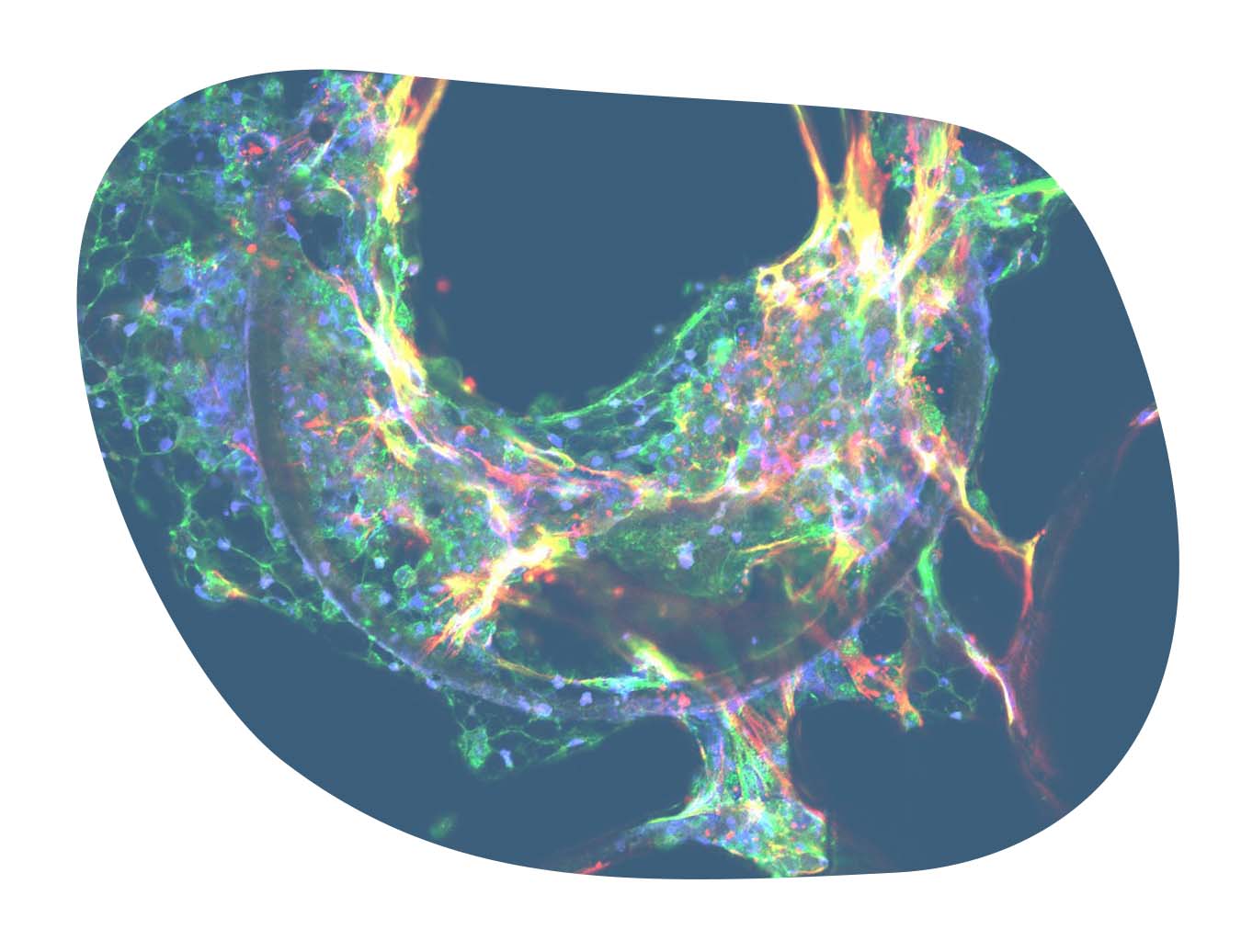
Recapitulating key aspects of human MASH disease
Our PhysioMimix MASH model captures key aspects of human disease including steatosis, inflammation, and fibrosis. By assessing biomarker expression, the assay provides high-content quantitative data from every replicate, including an assessment of fibrosis.
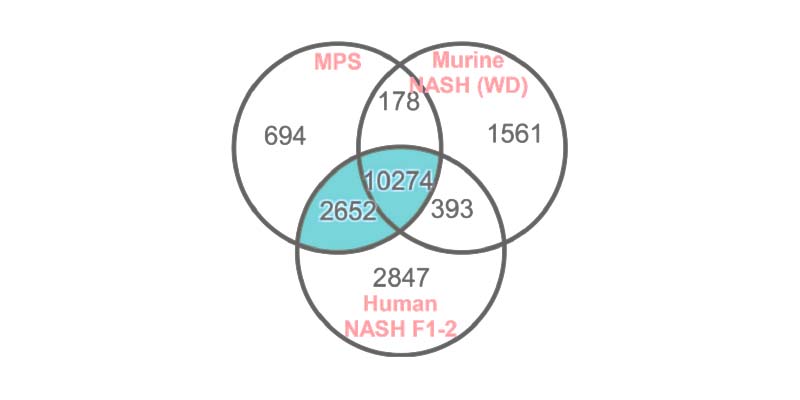
Improved alignment of transcriptomic profiles to human
Transcriptomic profiling of the PhysioMimix MASH model demonstrates a strong MASH profile when compared to human data and more closely replicates changes found in MASH patients than the murine WD model (Vacca et al., 2020).
A viable path forward for new drug modalities
The human-specific nature of new drug modalities exacerbates the interspecies challenges of using in vivo models to evaluate the efficacy of drugs to treat complex metabolic disorders during drug development. This siRNA study by Rao S et al., (2021) utilized the PhysioMimix MASH assay to explore the effect of siSPTBN1 treatment to prevent cancer development in diet-induced liver disease.
Access our MASLD/MASH Service
Get instant access to the PhysioMimix MASLD/MASH Assay via our CRO Service. Through a collaborative approach, our experts work with you to plan and execute your study.
Standard and bespoke projects are carried out by our dedicated team of scientists in our CRO facility providing you with actionable data within weeks.
PhysioMimix NASH-in-a-box kit
NASH-in-a-box contains everything that you require to recreate our PhysioMimix MASH Assay in your own laboratory.
Simply follow the software-guided protocol on your PhysioMimix Single- or Multi-organ microphysiological system to produce clinically translatable data fast.
Explore our Liver-on-a-chip models
Recreate the 3D multi-cellular architecture of the liver using perfused scaffolds. Achieve longer-term viability, enhanced functionality & high metabolic activity.
Frequently asked questions
What are the cell types in your human MASH in vitro model?
We use primary human hepatocytes, Kupffer cells and stellate cells with the ratio of 10:1:1.We source the primary cells from vendors including LifeNetHealth and Lonza, subject each lot to stringent validation testing to ensure assay performance in 3D tri-culture. More information about our validation process can be found in the following blog. To fast-track your route to assay adoption, we have developed a NASH-in-a-box kit for use with our PhysioMimix® OOC Systems. The kit contains everything that you need to recreate our human MASH in vitro assay in your laboratory.
What are the advantages of your human MASH in vitro model over spheroid models of fibrosis?
Spheroids are a very good system to study fibrosis, better than a 2D immortalised line model, however, spheroids models are still limited in terms of the number of end point measurements that can be detected, and their longevity.
The 3D tissue is totally perfused throughout an experiment, providing nutrients and O2 to all cells in the 3D tissue, thus enabling the culture of stable 3D tissue for longer periods of time (14 days for our standard NASH-in-a-box assay). Internally, we have run the assay for longer and offer flexibility in terms of assay duration through our MASH Contract Research Services. Contact us for more info.
Compared to static spheroid cultures, our human MASH in vitro model enables many more endpoints to be measured per well, due to the larger scale microtissue that we generate (over 480,000 cells per well) and large volume of media (~1 mL) from which to sample. Scaffolds containing the 3D liver microtissues can be cut in half; one half can be used for microscopy while the other can be used for -omics analysis e.g. RNAseq for transcriptomics. The large volume of media available for sampling allows for the analysis of various soluble biomarkers that include cell health (LDH, albumin, ALT/ AST), cell function (CYP activity), inflammation (IL-6, TNF-α, etc.) and fibrosis (TIMP1, Fibronectin)
Read the following publication for more info Kostrzewski et al, 2019.
Additionally, use of flow perfusion enables healthy (and diseased) liver tissue to remain functionally and metabolically active for over 4 weeks, much longer than is possible when culturing under static conditions. A co-publication with the FDA demonstrates the assay performance benefits of culturing using flow perfusion versus gold standard spheroid and liver sandwich approaches (Rubiano et al, 2020).
Have you tried any form of genetic modulation of the microtissues via CRISPR (for example) or via siRNA/ antisense?
We have used and adapted various genetic modulation methods in our system. We have looked at key single nucleotide polymorphisms (SNPs), for example, knocked-out patatin-like phospholipase domain containing 3 (PNPLA3) in Human Stellate cells (HSCs). In this study, we confirmed that mutated SNPs on PNPLA3 gene in HSC enhanced the overall MASH disease state (Kostrzewski et al, 2019). We have also successfully used small interfering RNA (siRNA) and antisense oligonucleotides (ASOs), both of which were shown to be effective in our model.
Have you tried to test for the progression of MASH into cancer or to combine your existing human MASH in vitro model with cancer cells?
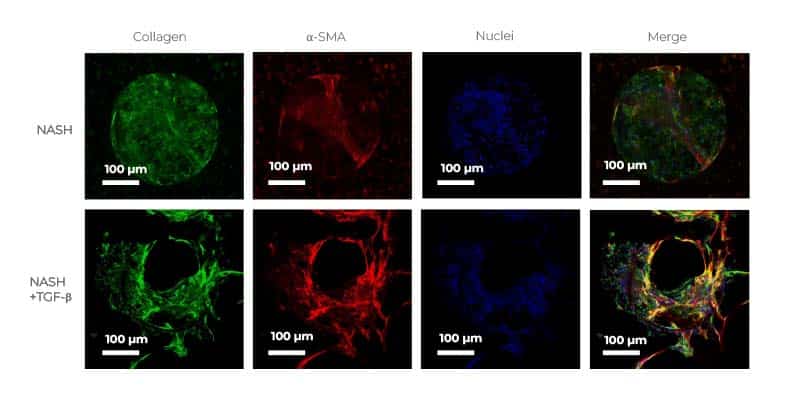
Fibrosis is key to the disease progression into severe MASH, cirrhosis and, if left untreated, liver cancer. Developing in vitro models that mimic the more advanced clinical phenotypes of the disease is therefore crucial, and it is something that we have recently started to investigate. For example, by adding cues such as tumor growth factor-beta (TGF-β), our 3D in vitro model can already progress to an advanced and more severe MASH stage with a quantifiable fibrosis phenotype (Kostrzewski et al, 2021), but we think we can go even further to reach a critical point where we generate, or trigger, hepatocellular cancer.
Have you tried iPS-derived hepatocytes instead of PHH in your human MASH in vitro model?
Our primary focus has always been to use primary cells to be as close to the human liver’s physiology as possible. However, there is a growing interest in developing iPS-derived hepatocytes models, especially for researchers who cannot easily access primary cells. We have successfully developed a monoculture liver model with iPS-derived hepatocytes, however this model has yet to be used to replicate a disease state or be used in a co-culture with other nonparenchymal cells (NPCs)
What end point analysis can be performed using your human MASH in vitro model?
We typically examine expression of various disease markers for steatosis (Nile Red), inflammation (IL-6, IL-8, TNF-α), and fibrosis (TIMP-1, Pro-collagen, Fibronectin, smooth muscle actin). Also, we evaluate cell function and health by measuring Urea, Albumin, LDH and Aspartate Transferase (AST)/Alanine amino transferase (ALT)
For deeper insights, periodic media sampling over time enables secreted biomarker assessments and post assay, RNA isolation for transcriptomic analysis.
Could you comment on the reproducibility and reliability of the PhysioMimix® OOC System and human MASH in vitro model?
As a rule, reliability is good between different batches. One of the advantages of CN Bio’s system is the ability to run a lot of experiments at the same time. We have 12 wells available on a single Multi-chip Liver-12 plate and we can run up to 6 plates at the same time. Generally, we get great reproducibility using the same donor within an experiment. When you start using different donors you do start to see some variability, but outcomes tend to trend the same. You would, however, expect to see donor to donor variations in the same way as variations are observed patient to patient. All cells used within the model are subjected to significant validation checks prior to experiments to ensure they give the best quality data. The following blog gives more information about our thorough lot validation process.
Can the human MASH in vitro model be used to understand key mechanistic data?
Please refer to the work of Vacca et al., 2020 who also explored TGFβ signalling and BMP signalling for modeling MASH. For BMP, their investigations confirmed that SMAD phosphorylation occurs. After 5 hours the target genes of SMAD are upregulated as you would expect. Vacca et al we were able to inhibit both the branches of the family using chemical inhibitors thus affecting the transduction of the signal. Their results concluded that the assay represents an effective in vitro tool for modeling MASH in preclinical science.
Additionally, our co-publication with AstraZeneca (Kostrzewski et al, 2021) explores transcriptional profile of MASH liver MPS model demonstrating that it aligns with profile from human clinical samples and the model’s potential for elucidating mechanism of action data. The scale of the MASH in vitro model is ideally suited to the analysis of multiple factors, both from the media and from the cells, allowing for transcriptome and proteomic analysis from every replicate.
Does the human MASH in vitro model represent a genuine alternative to animal models?
This topic is covered in more detail in the following blog.
How long can the human MASH in vitro model be cultured for?
As part of our R&D work, we have run MASH cultures for up to one month, providing plenty of time to include fat loading, genetic manipulation, and compound dosing. However, our NASH-in-a-box kit, which contains everything required to recreate our industry proven assay in your laboratory using a PhysioMimix® OOC System, is validated for use up to 14 days.
How flexible is the human MASH in vitro model, can it be customised?
Our model has been designed to be highly adaptable allowing for the culture of a range of primary cells. Our standard model, as utilized in our NASH-in-a-box kit, uses a tri-culture of hepatocytes, Kupffer cells and stellate cells with a modified media to support all three cell types. We can run simpler versions without non-parenchymal cells, however, there is the potential to develop more complex models too.
What throughput can be achieved, or how many conditions can be evaluated in one experiment using the human MASH in vitro model?
One PhysioMimix® OOC Controller unit can operate up to six Multi-chip Liver-12 plates at once with 12 wells in each plate. This results in a total throughput of 72 individual samples/run. As standard we run experiments in triplicate providing the ability to investigate 24 independent conditions. More complex studies can be scaled up using more than one PhysioMimix® Controller unit.
When using our NASH-in-a-box kit, two plates are supplied which means up to 6 conditions in triplicate can be run per kit.
What are your future plans for the human MASH in vitro model? Are there any plan to increase the scale?
Compared “like for like” versus technologies that use microfluidic flow (pneumatics that permit organ-specific tuning) instead of gravity-driven flow, we provide a higher throughput. Increasing the throughput is part of our roadmap now that we have the PhysioMimix® Higher Throughput (HT) System and Multi-chip Liver-48 plate, however, a commercialized solution is pending until a thorough evaluation of assay performance (in this miniaturized format) has been performed by our Science and Technology Team.
Where do you source your human primary cells from for your human MASH in vitro model? Have you used different donors? Do you see much variation in the level of fibrosis induction between donors?
When developing in vitro models, each cell lot/donor will react differently to the environment/conditions used. Some may do really well in 2D, under static conditions (e.g.: monolayer, liver sandwich or spheroid) but perform poorly under flow in a microphysiological system (MPS), or vice-versa. To develop the best model for your research, it is therefore important to source and test a variety of donors. A recent blog outlines the process that we go through to validate tri-cultures for MASH assay use.
We source our cells from a wide range of large cell suppliers such as Lonza, and LifeNet Health and thoroughly test each cell lot/donor to identify the best donors for our disease models. To save time and effort, customers can access CN Bio validated Hepatocyte and Kupffer cell lots (i.e. those that have been internally tested and proven to generate healthy, functional 3D liver tissues) through our 3D validated cell portfolio or directly from LifeNet Health. However, our NASH-in-a-box kit offers the fastest and easiest route to assay adoption, containing everything you need to recreate our model in your laboratory using PhysioMimix OOC.
Although using the same donor each time to generate data with consistency is ideal, it is impossible to guarantee as stocks of donors will run out at some point. It is also important to observe donor-donor variability to gain a better understanding of variations in metabolism between patients. When starting a project, we tend to use the same donor for a specific set of experiments to ensure consistency and build confidence in the data, then, switch to a different donor(s), to assess donor-donor variability. Donor variability can often be observed in our model; however, we generally see a consistent trend between donors. For example, when inducing severe fibrosis (by adding cues such as TGF-β), the severity of fibrosis may vary between donors, however, an increase in fibrosis is always observed.
What combination of cues gives the most MASH-like phenotype in your human MASH in vitro model?
When comparing cues/components, we look at expression profile using transcriptomics/RNA seq and compare the data to that of MASH patients to define the overlap. Although all cues are very good and show great MASH-like phenotype, TGF-β drives the MASH phenotype a little bit further than the other components. Depending on what researchers want to do with the model, adding different cues to subtly change the profile is possible. For example, if an inflammatory response is needed, lipopolysaccharide (LPS) is a very good component to add. Which cue to add and at what dose greatly depends on the research question being asked.
How does your OOC/MPS model compare to murine models?
This topic is covered in detail in the following blog.
If you do not find the answer to your question listed, please contact us