The human lung is in crisis
Our lungs represent the only internal organ that is directly subject to the environment and therefore face a relentless battle against foreign matter. For centuries, they have been assaulted by enemies – both natural and synthetic. Biological intruders such as viruses, bacteria and fungi constantly test defences that have evolved over thousands of years, whilst newer, stranger particles now enter the lung on an increasingly frequent basis, originating from burning fuels, disintegrating plastics and harmful chemicals made by our own human hands.
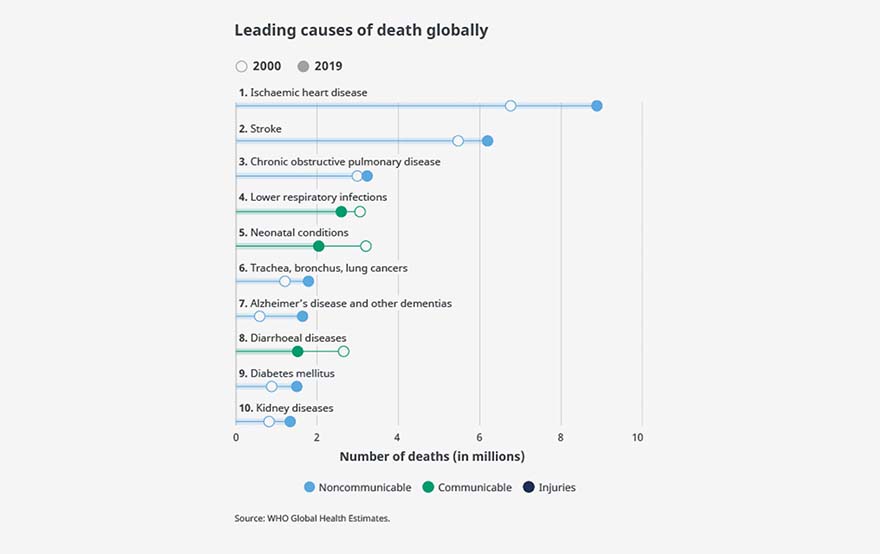
As a result, lung diseases have become one of the biggest causes of death; with chronic obstructive pulmonary disease (COPD), lower respiratory infections and lung cancer making up three of the top ten causes worldwide (World Health Organization (WHO), 2020). Unfortunately, the recent COVID-19 pandemic has further exacerbated this global crisis. As of March 2022, there has been almost 450 million confirmed cases of COVID-19 and 6 million deaths as reported by the WHO (WHO, 2022).
Figure 1. Leading causes of death globally – WHO Global Health Estimates.
Luckily for us, the resilient lung is well defended against encountered dangers. Specialised cells called Goblet cells produce a layer of gel-like mucus which coats the airway epithelium. Any larger particulate matter that enters the airway will become entrapped in the mucus. Then, via the “beat” of the cilia – a brush-like structure formed by ~50% of airway cells – the mucus is moved back up the airway and into the digestive system to be destroyed. This cycling of mucus – or mucociliary clearance – stops many, but not all, attacks to the lung.
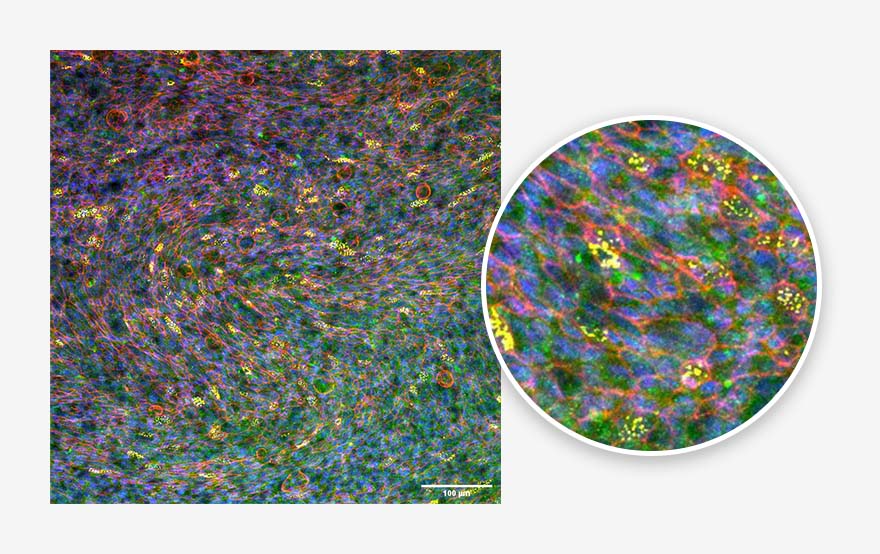
Figure 2. Bronchial epithelium. Tissue generated using the CN Bio PhysioMimix® OOC and Barrier (MPS-T12) Plates. Ciliated cells (yellow), mucus (green), actin cytoskeleton (red) and nuclei (blue) are indicated.
Cunning invaders that pass through frontline defences to reach the alveoli are only 0.2 μm away from reaching the bloodstream. However, the first immune sentinel of the lung – the alveolar macrophage – stands in their way. Alveolar macrophages patrol the internal portion of the alveoli and control the switch between anti-inflammatory tolerance and inflammatory attack response to foreign bodies. Intense inflammatory responses result in infiltration of circulating immune cells from the systemic blood whose aim is to destroy the invader but, unfortunately, this often causes damage to the alveoli too. In short, the wrong pathogen combined with inflammatory signalling from alveolar macrophages can represent the difference between a normal day and a trip to the emergency room.
This functional diversity, orchestrated by a multitude of pulmonary and immune cells, has proven challenging for scientists to reproduce in the laboratory. Traditional in vitro models range from simple 2D cell line cultures to 3D spheroids and scaffold models. Despite significant efforts and much improvement over the years, standard approaches fail to replicate many of the lung’s complex interactions
To overcome these shortfalls, animal models are widely used for lung research, despite differences in physiology and biology compared to human lung (Muñoz-Fontela et al., 2020; Sarkar & Heise, 2019). This disparity manifests in comparatively high drug attrition rates, with only 3% of pulmonary therapeutics reaching the market compared to 6-14% of therapeutics for other diseases (Barnes et al., 2015). Ex vivo studies, which utilise isolated, perfused lungs or tissue slices from human, represent an option to reduce animal experimentation and bring more physiological relevance to studies (Hocke et al., 2017; Viana et al., 2021); however, whole human lung tissues can be difficult to obtain, therefore precision cut lung slices (PCLS) provide a more cost-effective alternative.
The pros and cons of traditional approaches are summarised in more detail in Table 1 below, but the lack of easily accessible, cost-effective, human-representative preclinical models has resulted in preclinical pipeline blocks and shocking drug attrition rates.
| Model | Advantages | Disadvantages |
| 2D insert cell culture |
|
|
| Spheroid/organoid culture |
|
|
| Scaffold cultures |
|
|
| Ex vivo |
|
|
| In vivo |
|
|
Table 1. The pros and cons of traditional preclinical lung models
With the elevated risk of new pathogens causing future global pandemics, plus the air that we breathe becoming pollutant laden from things we take for granted – such as vehicles, gas central heating, paints, cleaning fluids, solvents and even farms – more rapid production of vaccines and therapeutics will be required. After the COVID-19 pandemic, organisations have begun to understand this requirement, for example the Bill Gates Foundation and Wellcome Trust recently donated $300 million dollars to the Coalition for Epidemic Preparedness Innovations (CEPI) with the aim to cut the time required to develop new vaccines. The same preparedness should be applied to the preclinical pipeline, whereby the effect of new pathogens, or irritants can be rapidly understood, therapeutics designed and tested to predict their efficacy and safety in human before final release to clinical trials.
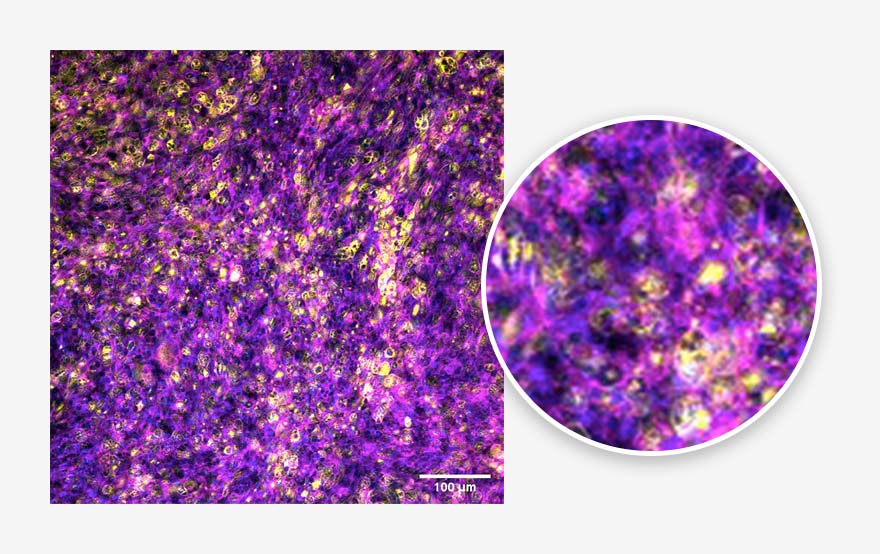
Figure 3. Alveolar epithelium. Tissue generated using the CN Bio PhysioMimix® OOC and Barrier (MPS-T12) Plates. ATII cells (yellow), actin cytoskeleton (magenta) and nuclei (blue) are indicated.
To meet this need, cost-effective, human-relevant models that faithfully recapitulate both healthy and diseased lung biology are urgently required. Lung-on-a-chip models (LOAC), alternatively known as lung microphysiological systems (MPS), have been in development over the last decade. Encouragingly, they have been shown to mimic aspects of the human lung better than other in vitro, thereby representing a potential solution.
A wide variety of LOAC models are now available, ranging from single-use microscale chips to insert-based multi-well plate systems that provide an easier way to incorporate immune cells and apply physiologically-relevant infection parameters (Artzy-Schnirman et al., 2019; Huh et al., 2010; Stucki et al., 2018; Zamprogno et al., 2021). LOAC models contain a combination of integrated microfluidics, scaffolds and stretchable membranes that expose cells to the shear stresses of the human lung, encouraging them to form microtissues that accurately mimic the human lung’s pathophysiology, as well as integrated sensors to monitor the tissues in real time. The more flexible solutions can be adapted to increase model complexity, via the co-culture of a wide range of cells, to address more specific research questions (Barkal et al., 2017; Humayun et al., 2018; Sellgren et al., 2014).
| Model | Advantages | Disadvantage |
| Organ-on-a-chip (OOC) or microphysiological systems (MPS) |
|
|
Table 2. The pros and cons of next generation LOAC.
Their ability to recreate physiologically relevant human lung tissue and monitor tissues in real time, means that LOAC models are well suited to studies that investigate infection and inflammatory responses to foreign matter (Benam et al., 2020; Deinhardt-Emmer et al., 2020; Zhang et al., 2020). Open-well format systems are particularly useful as test matter can be delivered into to the system as aerosols or small volumes at the ALI, and samples for analysis can be easily removed over time for longitudinal studies. The potential efficacy and safety of therapies in development can be tested via addition to the system apically at the ALI to simulate the dosing of inhaled medications, or basolaterally to mimic oral or intravenous therapies, whilst systemic effects can be elucidated by interconnecting relevant MPS organs together to form multi-organ systems (Chen et al., 2017; Edington et al., 2018). A summary of pros, cons and considerations when choosing an OOC solution can be found in Table 2.
Having read thus far, I hope you would agree that LOAC models offer much promise, but can they save the lung from crisis? Could they represent the missing piece of the preclinical puzzle? What if the complimentary use of LOAC models alongside traditional approaches eliminates our preclinical bottlenecks? Only time and further research will tell, but for now more you can learn more about LOAC models and their potential applications, at our next webinar:
Presented by Professor Wojciech Chrzanowski, Faculty of Medicine and Health, University of Sydney & Dr Emily Richardson, Lead Scientist – Assay Development, CN Bio on March 29th. We look forward to seeing you there!
Follow the links to learn more about CN Bio’s PhysioMimix OOC Systems and Lung-on-a-chip barrier models
AUTHOR
Dr Emily Richardson
Lead Scientist Assay Development – CN Bio
References:
Artzy-Schnirman, A., Zidan, H., Elias-Kirma, S., Ben-Porat, L., Tenenbaum-Katan, J., Carius, P., Fishler, R., Schneider-Daum, N., Lehr, C. M., & Sznitman, J. (2019). Capturing the onset of Bacterial Pulmonary Infection in Acini-on-Chips. Advanced Biosystems, 3(9), e1900026. https://doi.org/10.1002/ADBI.201900026
Barkal, L. J., Procknow, C. L., Álvarez-Garciá, Y. R., Niu, M., Jiménez-Torres, J. A., Brockman-Schneider, R. A., Gern, J. E., Denlinger, L. C., Theberge, A. B., Keller, N. P., Berthier, E., & Beebe, D. J. (2017). Microbial volatile communication in human organotypic lung models. Nature Communications 2017 8:1, 8(1), 1–10. https://doi.org/10.1038/s41467-017-01985-4
Barnes, P. J., Bonini, S., Seeger, W., Belvisi, M. G., Ward, B., & Holmes, A. (2015). Barriers to new drug development in respiratory disease. In European Respiratory Journal (Vol. 45, Issue 5, pp. 1197–1207). European Respiratory Society. https://doi.org/10.1183/09031936.00007915
Benam, K. H., Novak, R., Ferrante, T. C., Choe, Y., & Ingber, D. E. (2020). Biomimetic smoking robot for in vitro inhalation exposure compatible with microfluidic organ chips. Nature Protocols, 15(2), 183–206. https://doi.org/10.1038/S41596-019-0230-Y
Chen, W. L. K., Edington, C., Suter, E., Yu, J., Velazquez, J. J., Velazquez, J. G., Shockley, M., Large, E. M., Venkataramanan, R., Hughes, D. J., Stokes, C. L., Trumper, D. L., Carrier, R. L., Cirit, M., Griffith, L. G., & Lauffenburger, D. A. (2017). Integrated gut/liver microphysiological systems elucidates inflammatory inter-tissue crosstalk. Biotechnology and Bioengineering, 114(11), 2648–2659. https://doi.org/10.1002/BIT.26370
Deinhardt-Emmer, S., Rennert, K., Schicke, E., Cseresnyés, Z., Windolph, M., Nietzsche, S., Heller, R., Siwczak, F., Haupt, K. F., Carlstedt, S., Schacke, M., Figge, M. T., Ehrhardt, C., Löffler, B., & Mosig, A. S. (2020). Co-infection with Staphylococcus aureus after primary influenza virus infection leads to damage of the endothelium in a human alveolus-on-a-chip model. Biofabrication, 12(2). https://doi.org/10.1088/1758-5090/AB7073
Edington, C. D., Chen, W. L. K., Geishecker, E., Kassis, T., Soenksen, L. R., Bhushan, B. M., Freake, D., Kirschner, J., Maass, C., Tsamandouras, N., Valdez, J., Cook, C. D., Parent, T., Snyder, S., Yu, J., Suter, E., Shockley, M., Velazquez, J., Velazquez, J. J., … Griffith, L. G. (2018). Interconnected Microphysiological Systems for Quantitative Biology and Pharmacology Studies. Scientific Reports 2018 8:1, 8(1), 1–18. https://doi.org/10.1038/s41598-018-22749-0
Hocke, A. C., Suttorp, N., & Hippenstiel, S. (2017). Human lung ex vivo infection models. Cell and Tissue Research, 367(3), 511. https://doi.org/10.1007/S00441-016-2546-Z
Huh, D., Matthews, B. D., Mammoto, A., Montoya-Zavala, M., Yuan Hsin, H., & Ingber, D. E. (2010). Reconstituting organ-level lung functions on a chip. Science (New York, N.Y.), 328(5986), 1662–1668. https://doi.org/10.1126/SCIENCE.1188302
Humayun, M., Chow, C. W., & Young, E. W. K. (2018). Microfluidic lung airway-on-a-chip with arrayable suspended gels for studying epithelial and smooth muscle cell interactions. Lab on a Chip, 18(9), 1298–1309. https://doi.org/10.1039/C7LC01357D
Miller, A. J., & Spence, J. R. (2017). In vitro models to study human lung development, disease and homeostasis. Physiology, 32(3), 246–260. https://doi.org/10.1152/PHYSIOL.00041.2016/ASSET/IMAGES/LARGE/PHY0031703750002.JPEG
Muñoz-Fontela, C., Dowling, W. E., Funnell, S. G. P., Gsell, P. S., Riveros-Balta, A. X., Albrecht, R. A., Andersen, H., Baric, R. S., Carroll, M. W., Cavaleri, M., Qin, C., Crozier, I., Dallmeier, K., de Waal, L., de Wit, E., Delang, L., Dohm, E., Duprex, W. P., Falzarano, D., … Barouch, D. H. (2020). Animal models for COVID-19. Nature 2020 586:7830, 586(7830), 509–515. https://doi.org/10.1038/s41586-020-2787-6
Sarkar, S., & Heise, M. T. (2019). Mouse Models as Resources for Studying Infectious Diseases. Clinical Therapeutics, 41(10), 1912. https://doi.org/10.1016/J.CLINTHERA.2019.08.010
Sellgren, K. L., Butala, E. J., Gilmour, B. P., Randell, S. H., & Grego, S. (2014). A biomimetic multicellular model of the airways using primary human cells. Lab on a Chip, 14(17), 3349–3358. https://doi.org/10.1039/C4LC00552J
Stucki, J. D., Hobi, N., Galimov, A., Stucki, A. O., Schneider-Daum, N., Lehr, C. M., Huwer, H., Frick, M., Funke-Chambour, M., Geiser, T., & Guenat, O. T. (2018). Medium throughput breathing human primary cell alveolus-on-chip model. Scientific Reports, 8(1). https://doi.org/10.1038/S41598-018-32523-X
Viana, F., O’Kane, C. M., & Schroeder, G. N. (2021). Precision-cut lung slices: A powerful ex vivo model to investigate respiratory infectious diseases. Molecular Microbiology. https://doi.org/10.1111/MMI.14817
WHO. (2020). WHO Global Health Estimates. https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death
WHO. (2022). WHO Coronavirus (COVID-19) Dashboard . https://covid19.who.int/
Zamprogno, P., Wüthrich, S., Achenbach, S., Thoma, G., Stucki, J. D., Hobi, N., Schneider-Daum, N., Lehr, C. M., Huwer, H., Geiser, T., Schmid, R. A., & Guenat, O. T. (2021). Second-generation lung-on-a-chip with an array of stretchable alveoli made with a biological membrane. Communications Biology, 4(1). https://doi.org/10.1038/s42003-021-01695-0
Zhang, F., Liu, W., Zhou, S., Jiang, L., Wang, K., Wei, Y., Liu, A., Wei, W., & Liu, S. (2020). Investigation of Environmental Pollutant-Induced Lung Inflammation and Injury in a 3D Coculture-Based Microfluidic Pulmonary Alveolus System. Analytical Chemistry, 92(10), 7200–7208. https://doi.org/10.1021/ACS.ANALCHEM.0C00759/SUPPL_FILE/AC0C00759_SI_001.PDF