The liver is responsible for metabolizing most medications, and as such represents one of the primary tissues affected by adverse drug reactions. Drug-induced liver injury (DILI) is a potential complication of nearly all classes of medication. Unfortunately, it’s not at all uncommon for DILI to pass undetected during pre-clinical drug development or to be first identified in the clinic, where it can halt trials pending further investigations. In many cases, there’s little to worry about as, typically, ceasing the medication stops the DILI effect. However, it’s somewhat disconcerting that 750 FDA approved drugs are known to have at least some degree of DILI risk [Thakkar et al., 2017] and far too many potential new drugs are not approved due to an excessive DILI risk.
For attrition rates to fall, potential DILI liabilities in humans need to be identified earlier in the pre-clinical discovery pipeline. Doing so would let promising drugs return to the drawing board, where toxicity issues can potentially be “engineered out.” A number of challenges must be tackled when de-risking drug-induced liver injury, including:
- Animal DILI studies and their limited relevance to humans
-
- Animal models have a history of falsely identifying toxic drugs as safe and labeling useful drugs as toxic (Van Norman, 2019).
- For new human-specific drug modalities, animal physiology is unsuitable
- The complexity of standard in vitro test models needs to be improved such that they behave and respond to drugs in a way that more accurately represents the human body
- It has been historically challenging to use traditional approaches to screen idiosyncratic responses caused by pre-existing liver disease or genetic susceptibility.
The inconsistencies of animal models, paired with the limiting simplicity of 2D liver cultures, have led to a pressing need for more predictive pre-clinical in vitro tools to investigate DILI. Recent focus has centered on 3D cell culture platforms. Such approaches yield complex and functional human in vitro liver models that have the potential to overcome these challenges and guide more informed decision making when taking a drug from development to market [Weaver et al, 2020].
Fig 1: DILI is a multi-factorial pathogenesis
The in vitro DILI testing workflow
Every potential drug passes through a cascade of tests in early discovery, starting with simplistic high throughput batch screens for cell viability that ask the question – does the drug kill hepatocytes grown in 2D? As development progresses through the preclinical stage, there are far fewer drugs and concentrations to test. This reduction facilitates the use of lower throughput, more complex 3D tissue models. Through their adoption, researchers can more accurately predict the risk of DILI in vivo before entering the clinic.
The liver is a 3D tissue, consequently, 3D hepatocyte models, like cell spheroids, demonstrate improved results compared to 2D. Here, cells form physiologically relevant cellular interactions which ultimately yield more in vivo-like morphologies, metabolic activity, and extended culture longevity. In terms of assaying for DILI, batch screens in 2D typically pick up about a third of drugs that have adverse effects on the liver, however, a collaborative study between InSphero and AstraZeneca, using 3D primary human hepatocyte and non-parenchymal cell spheroids, improved upon this by identifying up to 60% of DILI positive compounds (Procter et al., 2017). But 40% remain undetected, so what kind of advancements beyond just 3D can help identify all drugs with DILI risk?
While some spheroid methods can result in hepatocyte polarization, these models generally fall short of the tissue-level structure seen in the liver. Furthermore, hepatocytes are exceptionally metabolic and require ample oxygen delivery, presenting challenges for spheroid cultures that are notoriously limited in nutrient diffusion. Without recapitulating high-order aspects of the liver, toxic drugs with subtle or very specific actions may pass through without notice. In conjunction with this, there are variables outside of the cell culture model that can influence DILI. Other organs and tissues can modulate how hepatocytes metabolize drugs. For instance, the kinetics of drug absorption by the gut can impact metabolite-driven toxicity in the liver. Along the same lines, disease pathophysiology (i.e. non-alcoholic fatty liver disorder (NAFLD)) can also change how the liver reacts to a drug, potentially exacerbating injury from smaller doses. Being able to incorporate these compounding factors into the pre-clinical toolbox would go a long way in ensuring only drugs with a low risk of liver injury move into clinical trials.
Gaining more relevant information through ‘flow’
The limitations mentioned above can be addressed by incorporating perfusion in a 3D culture. By growing physiologically relevant combinations of primary liver cells (including those with genetic variants that pre-dispose patients to disease) on continually perfused 3D scaffolds, the microarchitecture of the liver tissue can be closely reproduced. With fluid continually circulating to provide mechanical stimulus, consistent oxygen delivery, and polarization, hepatocytes in these microenvironments can maintain high functionality for up to 4 weeks.
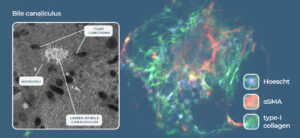
Fig 2: Liver-on-a-Chip models recreate the liver microarchitecture and can recapitulate the phenotype of metabolic diseases such as NASH
In addition to extending the realms of chronic dosing studies in vitro, these perfused Organ-on-a-Chip models (OOCs) offer greater flexibility to uncover immune-related toxicity issues by incorporating circulating immune cells into the system. It’s also possible to induce common liver disorders (such as NAFLD & Non-Alcoholic Steatohepatitis (NASH)) to unlock underlying morbidity susceptibilities.
To account for multi-organ effects on the toxicity of a compound, flow enables individual OOCs to be linked together into multi-organ systems. These systems can simulate essential processes such as drug absorption and metabolism and enable understanding of interactions between organs such as metabolite driven toxicity, multi-organ toxicity, or inflammation that may not have been apparent otherwise. Whilst their throughput may be lower, OOCs enable researchers to go beyond simply identifying the presence of acute/chronic liver toxicity or translating in vitro results into a clinical setting; they enable us to investigate the cause.
Answering more questions with multiple endpoints
While simpler systems successfully flag the presence or absence of a liver toxicity signal earlier than ever before in drug discovery, these are perfectly complimented by the use of more complex perfused OOCs to provide the detail. Achieving data richness and high assay sensitivity requires volume. Half a million primary cells perfused by large volumes of media (ranging between 1-2 mL) are required to generate enough horsepower to reproduce phase 1 and 2 metabolism (particularly important because one of the reasons that standard animal and in vitro models have proven to be poor predictors of DILI is due to their non-human metabolic profiles) and to increase assay sensitivity such that more sophisticated clinical chemistry outputs of liver function such as ALT/AST can be detected.
But this is worth the investment as scientists can now uncover unique insights not previously possible in vitro using simpler 2D or 3D systems, or even perfused Microphysiological Systems (MPS) that operate on a microscale using ”chip” technology. Samples taken over time in longitudinal studies provide biochemical assessments that build up a picture over time. The relatively large amount of tissue recovered at the end of the experiment can be examined in additional proteomics or genomics studies to look at gene up- and down-regulation to inform a more complete toxicology profile.
This ability to multiplex enables researchers to go beyond simply identifying the presence of acute or chronic liver toxicity, or to translate the results gained in vitro using lab grown organs into a clinical setting, but it enables us to investigate the mechanism behind the cause (Novac et al., 2021).

Understanding the cause enables solutions
Understanding the mechanism behind any given toxicity issue is invaluable. Such information allows the team an opportunity to reconfigure, potentially rescuing a project from failure. This iterative process is made significantly more efficient with a mechanistic fingerprint or a toxicity profile to inform the redesign.
Similarly, biological insights can help researchers determine what data is most relevant. From time to time, liver toxicology signals arise in pre-clinical animal testing that contradict clean human assays, or other in vivo animal species. Generating comparative human data before the first in-human study provides the confidence to proceed with reassurance that the signal is likely a function of the test animal species and not the compound itself. This knowledge may prevent the project from needlessly being terminated. Who knows how many potential blockbuster drugs have been prevented from achieving their potential? A short webinar from customers at the University of California -San Diego (UCSD) illustrating this very point.
Gaining toxicology insights beyond small molecules
Traditional toxicology work has tended to focus on small-molecule compounds, however, new human-specific modalities (like biologics, products encapsulated in lipid nanoparticles, or oligonucleotide/RNA-based therapies) have come to the fore of discovery and development. Although less prone to DILI, these new products still have the potential to induce liver injury. The Astellas-Audentes gene therapy trial for myotubular myopathy serves as a dire example of this. While the therapy yielded promising results, the death of 4 patients from liver failure has certainly called its future into question. In such a scenario, OOC is especially beneficial for testing safety since it eliminates the roadblocks of genetics, metabolism, and/or immunological responses that render animal models unsuitable while also addressing the lack of physiological flow and architecture of in vitro platforms. Indeed, OOCs are ‘agnostic’ to drug type and ensure the relevant receptors in the relevant parts of the tissue take up relevant molecules.
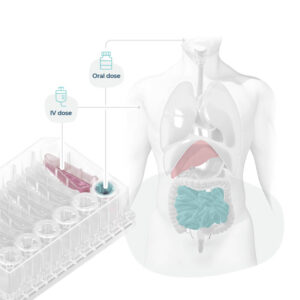
Fig 3: Organ-on-a-chips mimic organs phenotypes, enabling scientists to recreate human-relevant drug dosing and associated toxicity
Range of approaches
OOC enables deep toxicological insights that were not previously possible. An established consensus about OOC’s effectiveness, along with keen regulatory interest and political pressure to reduce animal usage in drug discovery, is driving adoption. Embedding OOC throughout drug discovery to uncover safety toxicology issues early will undoubtedly reduce risk and increase efficiency. Pharmaceutical and biotech companies are pursuing OOC technologies; larger companies may find establishing protocols in-house to be the best route whilst for smaller, or virtual biotech firms, full-service support from experts potentially offers a more practical approach. It is also important to note that, while different OOC systems are similar in some ways, they can be drastically different in others. As such, it is important to pick the right OOC for the job. As discussed above, it is crucial for DILI research to choose a system that: 1.) is easy to operate, 2.) yields large amounts of biological materials for sampling, 3.) incorporates physiologically relevant flow, and 3.) is ‘agnostic’ to therapy modality.
At CN Bio, we offer a range of OOC solutions. For those wishing to outsource, we offer a complete Drug Metabolism and Safety Toxicity Testing Service, whilst in-house adoption is facilitated by the PhysioMimix Core microphysiological systems, and DILI assay kit: Human 24. Our system utilizes an open-top plate that is familiar to most cell culture scientists and is capable of producing large amounts of sample (both cell number and media) while maintaining continuous flow to generate physiologically relevant results.
Watch this on-demand webinar discussing how a human Liver-on-a-chip, otherwise referred to as a liver microphysiological system, can be used to understand the causality and mechanistic aspects of drug-induced liver injury (DILI). Using a broad set of mild to severe hepatotoxic compounds the MPS predicted the toxicity of drugs that had not previously been identified in vitro.
AUTHOR
Dr Anthony Berger
Field Application Scientist, CN Bio
AUTHOR