The aim of organ-on-a-chip (OOC) technology is to more accurately replicate human physiology in vitro to overcome the relevance limitations of current approaches, and the field is progressing at a rapid pace! But are they animal alternatives?
Many hundreds of academic researchers are choosing to work in this area, supported by an increasing number of grant funding opportunities. There is exponential growth in the number of publications that cite the use of, or development of, OOC technologies and a collection of well-established companies that design and produce commercially viable systems and services. Major Pharma and Biotech are showing an appetite to help bring new medicines to market more efficiently and, importantly, there is genuine interest in OOC from the regulators1.
Whilst these advanced OOC models clearly improve physiological relevance and culture longevity versus standard 2D cell cultures, do they compare so favourably versus in vivo models? We are frequently asked; will OOC replace the use of animals and, show me data to prove your model is better than in vivo models? Whilst ethically desirable and certainly an aim for most people working in the organ-on-a-chip field, realistically, replacing animal usage completely is unlikely to happen for quite some time. Before this aim can become a reality there is still much to prove, however, by incorporating OOC into drug discovery and development at strategic stages, these disruptive technologies can be used to cross-validate and supplement data sets right now! So, perhaps a more pertinent question to ask is, what are the disadvantages of animal models and how can OOC be used in combination to fill knowledge gaps?
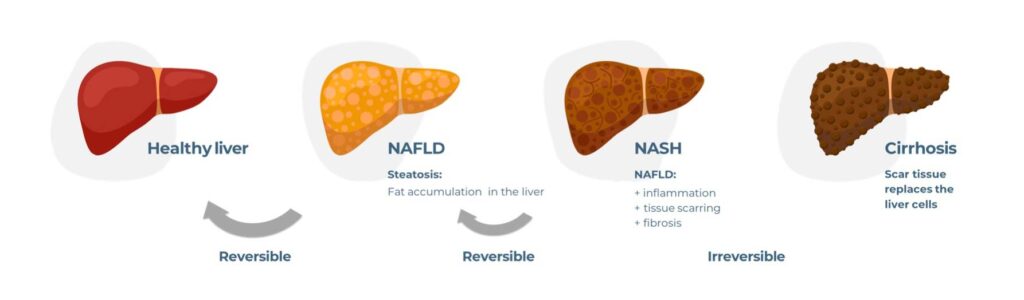
Using the field of liver research as an example, we will explore this concept further. The most common cause of chronic liver disease worldwide is now a metabolic disorder called Non-alcoholic fatty liver disease (NAFLD – also known as Metabolic dysfunction-associated steatotic liver disease MASLD)), which is linked to both obesity and diabetes and affects a staggering 25% of the global population. NAFLD is a spectrum of metabolic diseases in which fat builds up in the liver, due to diet and lifestyle, in the absence of excess alcohol consumption. Approximately 20% of people diagnosed with NAFLD go on to develop Non-alcoholic steatohepatitis (NASH – also known as Metabolic dysfunction-associated steatohepatitis (MASH)), which is associated with steatosis (fat deposits in the liver), inflammation, fibrosis, and insulin resistance. Left untreated, NASH can progress further into cirrhosis and/or Hepatocellular carcinoma (HCC), for which a liver transplant is the only possible therapeutic intervention. To prevent NASH from becoming a global economic burden, many companies have embarked on drug discovery programmes, yet their high-profile compounds continually fail to meet primary endpoints in late-stage clinical trials. To this day NASH remains a disease of unmet medical need.
What are the disadvantages of pre-clinical in vivo models of NASH?
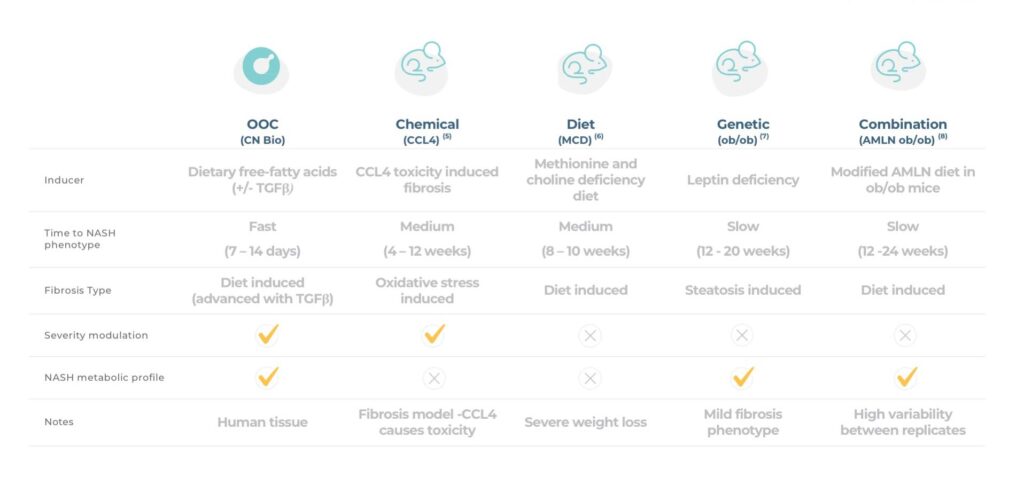
Despite there being a wide variety of pre-clinical NASH models available, predicting the efficacy of developmental drugs in humans remains a challenge. While in vivo models offer a clear “systems” advantage over current in vitro models, they are, at the end of the day, mice and not humans, so cross-species differences remain a valid data translatability concern. In vivo murine models are by far the most widely used pre-clinical tool and are generally grouped into the following categories: genetic, dietary, chemical and combinations thereof. Unfortunately, there’s no easy “one size fits all” option.
1. Genetic
Genetic mouse models are useful for studying specific aspects of NASH including obesity and HCC. Starting with the former, Ob/ob mice deficient in leptin, and db/db mice with non-functioning leptin receptors, are used to model obesity leading to severe steatosis – a hallmark of NASH. Whilst these models are brilliant at generating fat mice, unfortunately they don’t replicate other key markers of NASH, such as hepatocellular ballooning and inflammation. To produce a more representative NASH phenotype, leptin models need to be combined with dietary or chemical elements.
Likewise, a combination of genetic (e.g., tumour suppressor gene PTEN mutations) and dietary models is required to investigate the predisposition of NASH patients to HCC2.
2. Dietary
Much like genetic models, dietary models capture certain elements of NASH but fail to recapitulate the entire disease. To compensate for their limitations, dietary models are also frequently used in combination with genetic, or chemical elements. One of the most researched models of NAFLD, the methionine and choline-deficient (MCD) diet, is an excellent example. This diet is high in sugars and fats but deficient in choline and methionine. It results in the accumulation of fat in hepatocytes, oxidative stress, inflammation, ballooning and fibrosis, and causes increased alanine transaminase (ALT) and aspartate transaminase (AST), all key clinical markers of NASH. Unfortunately, the metabolic profile induced by this diet is very different to human NASH with weight loss observed rather than obesity, as well as an absence of insulin resistance and dyslipidaemia3.
In general, high-fat diets better recapitulate the overall NASH metabolic phenotype than MCD diets. Treated mice develop obesity, insulin resistance and hyperlipidaemia, but fall short of MCD diets for inducing steatosis, inflammation and fibrosis4.
3. Chemical
Generally, chemical models work best in combination with dietary and genetic models to address phenotype shortfalls. For example, Streptozotocin is used to induce diabetes and, when combined with a high-fat diet, can be used to replicate NAFLD, whilst Diethylnitrosamine can be added to a high-fat diet to induce HCC4. Carbon tetrachloride (CCl4) induces an oxidative stress response leading to hepatic fibrosis, however, on the negative side, it causes severe toxicity and, when combined with a high-fat diet, a reduction in body weight5. All in all, this makes the interpretation of experimental results a challenge.
One question remains unanswered in my mind too, how relevant to the mechanisms of disease onset in humans are chemically induced models and could that also have a bearing on a lack of a lack of data translatability?
I’m sure you get the picture; each mouse model can replicate some key aspects of NASH but not all. When models are combined, they can mimic the major disease hallmarks, but in doing so may cause “side effects” that are not typical to NASH. The requirement to combine or alter the models to suit specific research needs is complex, time-consuming, expensive, extremely difficult to validate and the data derived from them isn’t reliably translating into clinical outcomes.
How does Organ-on-a-chip complement existing disease models?
One thing is for sure, there’s little chance of pre-clinical drugs achieving clinical success if pre-clinical models lack human predictivity, unless blessed by a very lucky dose of serendipity. Going back to our NASH example, does this mean that we should give up on R&D efforts, or is there simply an important weapon missing in the arsenal? Also, what if you are developing drugs against targets that might not be expressed in murine models? Can Organ-on-a-chip help to plug the gaps to avoid quite such a muddle of models?
Our advanced in vitro model of NASH disease is cultured using the PhysioMimix® OOC. It utilises our proprietary liver-on-a-chip technology to provide long-term in vitro cultures of primary human hepatocytes, stellate and Kupffer cells in 3D microtissue structures that capture key NAFLD/NASH stages: intracellular fat accumulation, inflammation and fibrosis. The model enables the precise mechanistic effects of compounds to be teased out, and for models to be easily manipulated to suit research needs9. Co-authored research, led by the University of Cambridge in 202010, illustrates the direct translation between this NASH model, human clinical studies and in vivo mouse models. Their research demonstrates that this NASH model delivers more human-relevant data than standard in vitro liver tissue studies, it represents a quicker and cheaper option to study NASH pathophysiology than long and expensive animal models, whilst providing similar/confirmatory results. Data generated using this NASH model helped to elucidate the complex cellular mechanisms that cause liver fibrosis, identifying a novel pathway that regulates the underlying mechanism of NASH, and potentially a new therapeutic target. The results speak for themselves, OOC is most certainly a tool available today to help reduce, refine, and complement existing tests. Where mice capture the complexity of a full organism, OOC models demonstrate how a disease mechanism, or the effects of a drug will differ in a human setting. The combination of the two gives a much stronger and broader insight that aids translational research. However, should you be focussed on targets that are human-specific, where the use of animal models is less suited, OOC can be used as an alternative right now!
So, as we look to the future we remain hopeful that the complimentary, or alternative use of OOC models will help to pave the way for a successful (NASH) therapeutic to make it to market. In December 2023, we were delighted to announce that our PhysioMimix NASH assay was used to provide human-relevant data on compound efficacy to support the initiation of Inipharm’s Phase 1 clinical trial for INI-822. The submission represents the first example of an OOC provider’s data supporting the clinical progression of a drug for a complex metabolic liver disease and demonstrates the transformative potential of these models to provide human-relevant insights within preclinical programs.
Plus, as these advanced in vitro models continue to increase in sophistication from individual OOC models that operate in isolation into multi-organ models that start to represent systemic functions and beyond, the future vision is more firmly focused on reducing and replacing our reliance on animal models where improved performance is proven.
Adopting our NASH model into your workflow
We have developed a unique product called NASH in-a-box, which is tailored for use with our PhysioMimix® OOC range of microphysiological systems. The kit includes all the primary human cells, media and consumables required to recreate our NASH model in your laboratory. Combined with software-guided protocols (which walk you through the experimental process step-by-step), the kit streamlines the entire workflow, enabling rapid adoption. Alternatively, you can access the model through our portfolio of Contract Research Services. More information about our PhysioMimix NASH assay can be found here.
Blog updated in 2024.
AUTHOR
Gareth Guenigault
Senior Scientist, CN Bio
To learn more watch the following webinar on demand:
- Rubiano, A., Indapurkar, A., Yokosawa, R., Miedzik, A., Rosenzweig, B., Arefin, A., Moulin, C.M., Dame, K., Hartman, N., Volpe, D.A., Matta, M.K., Hughes, D.J., Strauss, D.G., Kostrzewski, T. and Ribeiro, A.J.S. (2021), Characterizing the reproducibility in using a liver microphysiological system for assaying drug toxicity, metabolism, and accumulation. Clin Transl Sci, 14: 1049-1061. https://doi.org/10.1111/cts.12969
- Horie Y, Suzuki A, Kataoka E, Sasaki T, Hamada K, Sasaki J, Mizuno K, Hasegawa G, Kishimoto H, Iizuka M, Naito M, Enomoto K, Watanabe S, Mak TW, Nakano T. Hepatocyte-specific Pten deficiency results in steatohepatitis and hepatocellular carcinomas. J Clin Invest. 2004 Jun;113(12):1774-83. doi: 10.1172/JCI20513. PMID: 15199412; PMCID: PMC420505.
- Ibrahim SH, Hirsova P, Malhi H, Gores GJ. Animal Models of Nonalcoholic Steatohepatitis: Eat, Delete, and Inflame. Dig Dis Sci. 2016 May;61(5):1325-36. doi: 10.1007/s10620-015-3977-1. Epub 2015 Dec 1. PMID: 26626909; PMCID: PMC4838538.
- Van Herck MA, Vonghia L, Francque SM. Animal Models of Nonalcoholic Fatty Liver Disease-A Starter’s Guide. Nutrients. 2017 Sep 27;9(10):1072. doi: 10.3390/nu9101072. PMID: 28953222; PMCID: PMC5691689.
- Scholten D, Trebicka J, Liedtke C, Weiskirchen R. The carbon tetrachloride model in mice. Lab Anim. 2015 Apr;49(1 Suppl):4-11. doi: 10.1177/0023677215571192. PMID: 25835733.
- Takahashi Y, Soejima Y, Fukusato T. Animal models of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. World J Gastroenterol. 2012;18(19):2300-2308. doi:10.3748/wjg.v18.i19.2300
- Trak-Smayra V, Paradis V, Massart J, Nasser S, Jebara V, Fromenty B. Pathology of the liver in obese and diabetic ob/ob and db/db mice fed a standard or high-calorie diet. Int J Exp Pathol. 2011 Dec;92(6):413-21. doi: 10.1111/j.1365-2613.2011.00793.x. Epub 2011 Nov 25. PMID: 22118645; PMCID: PMC3248077.
- Criver.com – Nonalcoholic Steatohepatitis NASH Models
- Kostrzewski T, Maraver P, Ouro-Gnao L, Levi A, Snow S, Miedzik A, Rombouts K, Hughes D. A Microphysiological System for Studying Nonalcoholic Steatohepatitis. Hepatol Commun. 2019 Nov 13;4(1):77-91. doi: 10.1002/hep4.1450. PMID: 31909357; PMCID: PMC6939502.
- Vacca M, Leslie J, Virtue S, Lam BYH, Govaere O, Tiniakos D, Snow S, Davies S, Petkevicius K, Tong Z, Peirce V, Nielsen MJ, Ament Z, Li W, Kostrzewski T, Leeming DJ, Ratziu V, Allison MED, Anstee QM, Griffin JL, Oakley F, Vidal-Puig A. Bone morphogenetic protein 8B promotes the progression of non-alcoholic steatohepatitis. Nat Metab. 2020 Jun;2(6):514-531. doi: 10.1038/s42255-020-0214-9. Epub 2020 Jun 8. PMID: 32694734.



